How old fishing nets turn into chairs, car mats and Prada bags

A company called Aquafil is turning the fibers from fishing nets and old carpets into new threads for chairs, car mats, bikinis and Prada bags.
Discarded nylon fishing nets in the oceans are among the most harmful forms of plastic pollution. Every year, about 640,000 tons of fishing gear are left in our oceans and other water bodies to turn into death traps for marine life. London-based non-profit World Animal Protection estimates that entanglement in this “ghost gear” kills at least 136,000 seals, sea lions and large whales every year. Experts are challenged to estimate how many birds, turtles, fish and other species meet the same fate because the numbers are so high.
Since 2009, Giulio Bonazzi, the son of a small textile producer in northern Italy, has been working on a solution: an efficient recycling process for nylon. As CEO and chairman of a company called Aquafil, Bonazzi is turning the fibers from fishing nets – and old carpets – into new threads for car mats, Adidas bikinis, environmentally friendly carpets and Prada bags.
For Bonazzi, shifting to recycled nylon was a question of survival for the family business. His parents founded a textile company in 1959 in a garage in Verona, Italy. Fifteen years later, they started Aquafil to produce nylon for making raincoats, an enterprise that led to factories on three continents. But before the turn of the century, cheap products from Asia flooded the market and destroyed Europe’s textile production. When Bonazzi had finished his business studies and prepared to take over the family company, he wondered how he could produce nylon, which is usually produced from petrochemicals, in a way that was both successful and ecologically sustainable.
The question led him on an intellectual journey as he read influential books by activists such as world-renowned marine biologist Sylvia Earle and got to know Michael Braungart, who helped develop the Cradle-to-Cradle ethos of a circular economy. But the challenges of applying these ideologies to his family business were steep. Although fishing nets have become a mainstay of environmental fashion ads—and giants like Dupont and BASF have made breakthroughs in recycling nylon—no one had been able to scale up these efforts.
For ten years, Bonazzi tinkered with ideas for a proprietary recycling process. “It’s incredibly difficult because these products are not made to be recycled,” Bonazzi says. One complication is the variety of materials used in older carpets. “They are made to be beautiful, to last, to be useful. We vastly underestimated the difficulty when we started.”
Soon it became clear to Bonazzi that he needed to change the entire production process. He found a way to disintegrate old fibers with heat and pull new strings from the discarded fishing nets and carpets. In 2022, his company Aquafil produced more than 45,000 tons of Econyl, which is 100% recycled nylon, from discarded waste.
More than half of Aquafil’s recyclate is from used goods. According to the company, the recycling saves 90 percent of the CO2 emissions compared to the production of conventional nylon. That amounts to saving 57,100 tons of CO2 equivalents for every 10,000 tons of Econyl produced.
Bonazzi collects fishing nets from all over the world, including Norway and Chile—which have the world’s largest salmon productions—in addition to the Mediterranean, Turkey, India, Japan, Thailand, the Philippines, Pakistan, and New Zealand. He counts the government leadership of Seychelles as his most recent client; the island has prohibited ships from throwing away their fishing nets, creating the demand for a reliable recycler. With nearly 3,000 employees, Aquafil operates almost 40 collection and production sites in a dozen countries, including four collection sites for old carpets in the U.S., located in California and Arizona.
First, the dirty nets are gathered, washed and dried. Bonazzi explains that nets often have been treated with antifouling agents such as copper oxide. “We recycle the coating separately,” he says via Zoom from his home near Verona. “Copper oxide is a useful substance, why throw it away?”
Still, only a small percentage of Aquafil’s products are made from nets fished out of the ocean, so your new bikini may not have saved a strangled baby dolphin. “Generally, nylon recycling is a good idea,” says Christian Schiller, the CEO of Cirplus, the largest global marketplace for recyclates and plastic waste. “But contrary to what consumers think, people rarely go out to the ocean to collect ghost nets. Most are old, discarded nets collected on land. There’s nothing wrong with this, but I find it a tad misleading to label the final products as made from ‘ocean plastic,’ prompting consumers to think they’re helping to clean the oceans by buying these products.”
Aquafil gets most of its nets from aqua farms. Surprisingly, one of Aquafil’s biggest problems is finding enough waste. “I know, it’s hard to believe because waste is everywhere,” Bonazzi says. “But we need to find it in reliable quantity and quality.” He has invested millions in establishing reliable logistics to source the fishing nets. Then the nets get shredded into granules that can be turned into car mats for the new Hyundai Ioniq 5 or a Gucci swimsuit.
The process works similarly with carpets. In the U.S. alone, 3.5 billion pounds of carpet are discarded in landfills every year, and less than 3 percent are currently recycled. Aquafil has built a recycling plant in Phoenix to help divert 12,500 tons of carpets from the landfill every year. The carpets are shredded and deconstructed into three components: fillers such as calcium carbonate will be reused in the cement industry, synthetic fibers like polypropylene can be used for engineering plastics, and nylon. Only the pelletized nylon gets shipped back to Europe for the production of Econyl. “We ship only what’s necessary,” Bonazzi says. Nearly 50 percent of his nylon in Italy and Slovenia is produced from recyclate, and he hopes to increase the percentage to two-thirds in the next two years.
His clients include Interface, the leading world pioneer for sustainable flooring, and many other carpet producers plus more than 2500 fashion labels, including Gucci, Prada, Patagonia, Louis Vuitton, Adidas and Stella McCartney. “Stella McCartney just introduced a parka that’s made 100 percent from Econyl,” Bonazzi says. “We’re also in a lot of sportswear because Nylon is a good fabric for swimwear and for yoga clothes.” Next, he’s looking into sunglasses and chairs made with Econyl - for instance, the flexible ergonomic noho chair, designed by New Zealand company Formway.
“When I look at a landfill, I see a gold mine," Bonazzi says.
“Bonazzi decided many years ago to invest in the production of recycled nylon though industry giants halted similar plans after losing large investments,” says Anika Herrmann, vice president of the German Greentech-competitor Camm Solutions, which creates bio-based polymers from cane sugar and other ag waste. “We need role models like Bonazzi who create sustainable solutions with courage and a pioneering spirit. Like Aquafil, we count on strategic partnerships to enable fast upscaling along the entire production chain.”
Bonazzi’s recycled nylon is still five to 10 percent more expensive than conventionally produced material. However, brands are increasingly bending to the pressure of eco-conscious consumers who demand sustainable fashion. What helped Bonazzi was the recent rise of oil prices and the pressure on industries to reduce their carbon footprint. Now Bonazzi says, “When I look at a landfill, I see a gold mine.”
Ideally, the manufacturers take the products back when the client is done with it, and because the nylon can theoretically be reused nearly infinitely, the chair or bikini could be made into another chair or bikini. “But honestly,” Bonazzi half-jokes, “if someone returns a McCartney parka to me, I’ll just resell it because it’s so expensive.”
The next step: Bonazzi wants to reshape the entire nylon industry by pivoting from post-consumer nylon to plant-based nylon. In 2017, he began producing “nylon-6,” together with Genomatica in San Diego. The process uses sugar instead of petroleum. “The idea is to make the very same molecule from sugar, not from oil,” he says. The demonstration plant in Ljubljana, Slovenia, has already produced several hundred tons of nylon, and Genomatica is collaborating with Lululemon to produce plant-based yoga wear.
Bonazzi acknowledges that his company needs a few more years before the technology is ready to meet his ultimate goal, producing only recyclable products with no petrochemicals, low emissions and zero waste on an industrial scale. “Recycling is not enough,” he says. “You also need to produce the primary material in a sustainable way, with a low carbon footprint.”
DNA gathered from animal poop helps protect wildlife
Alida de Flamingh and her team are collecting elephant dung. It holds a trove of information about animal health, diet and genetic diversity.
On the savannah near the Botswana-Zimbabwe border, elephants grazed contentedly. Nearby, postdoctoral researcher Alida de Flamingh watched and waited. As the herd moved away, she went into action, collecting samples of elephant dung that she and other wildlife conservationists would study in the months to come. She pulled on gloves, took a swab, and ran it all over the still-warm, round blob of elephant poop.
Sequencing DNA from fecal matter is a safe, non-invasive way to track and ultimately help protect over 42,000 species currently threatened by extinction. Scientists are using this DNA to gain insights into wildlife health, genetic diversity and even the broader environment. Applied to elephants, chimpanzees, toucans and other species, it helps scientists determine the genetic diversity of groups and linkages with other groups. Such analysis can show changes in rates of inbreeding. Populations with greater genetic diversity adapt better to changes and environmental stressors than those with less diversity, thus reducing their risks of extinction, explains de Flamingh, a postdoctoral researcher at the University of Illinois Urbana-Champaign.
Analyzing fecal DNA also reveals information about an animal’s diet and health, and even nearby flora that is eaten. That information gives scientists broader insights into the ecosystem, and the findings are informing conservation initiatives. Examples include restoring or maintaining genetic connections among groups, ensuring access to certain foraging areas or increasing diversity in captive breeding programs.
Approximately 27 percent of mammals and 28 percent of all assessed species are close to dying out. The IUCN Red List of threatened species, simply called the Red List, is the world’s most comprehensive record of animals’ risk of extinction status. The more information scientists gather, the better their chances of reducing those risks. In Africa, populations of vertebrates declined 69 percent between 1970 and 2022, according to the World Wildlife Fund (WWF).
“We put on sterile gloves and use a sterile swab to collect wet mucus and materials from the outside of the dung ball,” says Alida de Flamingh, a postdoctoral researcher at the University of Illinois Urbana-Champaign.
“When people talk about species, they often talk about ecosystems, but they often overlook genetic diversity,” says Christina Hvilsom, senior geneticist at the Copenhagen Zoo. “It’s easy to count (individuals) to assess whether the population size is increasing or decreasing, but diversity isn’t something we can see with our bare eyes. Yet, it’s actually the foundation for the species and populations.” DNA analysis can provide this critical information.
Assessing elephants’ health
“Africa’s elephant populations are facing unprecedented threats,” says de Flamingh, the postdoc, who has studied them since 2009. Challenges include ivory poaching, habitat destruction and smaller, more fragmented habitats that result in smaller mating pools with less genetic diversity. Additionally, de Flamingh studies the microbial communities living on and in elephants – their microbiomes – looking for parasites or dangerous microbes.
Approximately 415,000 elephants inhabit Africa today, but de Flamingh says the number would be four times higher without these challenges. The IUCN Red List reports African savannah elephants are endangered and African forest elephants are critically endangered. Elephants support ecosystem biodiversity by clearing paths that help other species travel. Their very footprints create small puddles that can host smaller organisms such as tadpoles. Elephants are often described as ecosystems’ engineers, so if they disappear, the rest of the ecosystem will suffer too.
There’s a process to collecting elephant feces. “We put on sterile gloves (which we change for each sample) and use a sterile swab to collect wet mucus and materials from the outside of the dung ball,” says de Flamingh. They rub a sample about the size of a U.S. quarter onto a paper card embedded with DNA preservation technology. Each card is air dried and stored in a packet of desiccant to prevent mold growth. This way, samples can be stored at room temperature indefinitely without the DNA degrading.
Earlier methods required collecting dung in bags, which needed either refrigeration or the addition of preservatives, or the riskier alternative of tranquilizing the animals before approaching them to draw blood samples. The ability to collect and sequence the DNA made things much easier and safer.
“Our research provides a way to assess elephant health without having to physically interact with elephants,” de Flamingh emphasizes. “We also keep track of the GPS coordinates of each sample so that we can create a map of the sampling locations,” she adds. That helps researchers correlate elephants’ health with geographic areas and their conditions.
Although de Flamingh works with elephants in the wild, the contributions of zoos in the United States and collaborations in South Africa (notably the late Professor Rudi van Aarde and the Conservation Ecology Research Unit at the University of Pretoria) were key in studying this method to ensure it worked, she points out.
Protecting chimpanzees
Genetic work with chimpanzees began about a decade ago. Hvilsom and her group at the Copenhagen Zoo analyzed DNA from nearly 1,000 fecal samples collected between 2003 and 2018 by a team of international researchers. The goal was to assess the status of the West African subspecies, which is critically endangered after rapid population declines. Of the four subspecies of chimpanzees, the West African subspecies is considered the most at-risk.
In total, the WWF estimates the numbers of chimpanzees inhabiting Africa’s forests and savannah woodlands at between 173,000 and 300,000. Poaching, disease and human-caused changes to their lands are their major risks.
By analyzing genetics obtained from fecal samples, Hvilsom estimated the chimpanzees’ population, ascertained their family relationships and mapped their migration routes.
“One of the threats is mining near the Nimba Mountains in Guinea,” a stronghold for the West African subspecies, Hvilsom says. The Nimba Mountains are a UNESCO World Heritage Site, but they are rich in iron ore, which is used to make the steel that is vital to the Asian construction boom. As she and colleagues wrote in a recent paper, “Many extractive industries are currently developing projects in chimpanzee habitat.”
Analyzing DNA allows researchers to identify individual chimpanzees more accurately than simply observing them, she says. Normally, field researchers would install cameras and manually inspect each picture to determine how many chimpanzees were in an area. But, Hvilsom says, “That’s very tricky. Chimpanzees move a lot and are fast, so it’s difficult to get clear pictures. Often, they find and destroy the cameras. Also, they live in large areas, so you need a lot of cameras.”
By analyzing genetics obtained from fecal samples, Hvilsom estimated the chimpanzees’ population, ascertained their family relationships and mapped their migration routes based upon DNA comparisons with other chimpanzee groups. The mining companies and builders are using this information to locate future roads where they won’t disrupt migration – a more effective solution than trying to build artificial corridors for wildlife.
“The current route cuts off communities of chimpanzees,” Hvilsom elaborates. That effectively prevents young adult chimps from joining other groups when the time comes, eventually reducing the currently-high levels of genetic diversity.
“The mining company helped pay for the genetics work,” Hvilsom says, “as part of its obligation to assess and monitor biodiversity and the effect of the mining in the area.”
Of 50 toucan subspecies, 11 are threatened or near-threatened with extinction because of deforestation and poaching.
Identifying toucan families
Feces aren't the only substance researchers draw DNA samples from. Jeffrey Coleman, a Ph.D. candidate at the University of Texas at Austin relies on blood tests for studying the genetic diversity of toucans---birds species native to Central America and nearby regions. They live in the jungles, where they hop among branches, snip fruit from trees, toss it in the air and catch it with their large beaks. “Toucans are beautiful, charismatic birds that are really important to the ecosystem,” says Coleman.
Of their 50 subspecies, 11 are threatened or near-threatened with extinction because of deforestation and poaching. “When people see these aesthetically pleasing birds, they’re motivated to care about conservation practices,” he points out.
Coleman works with the Dallas World Aquarium and its partner zoos to analyze DNA from blood draws, using it to identify which toucans are related and how closely. His goal is to use science to improve the genetic diversity among toucan offspring.
Specifically, he’s looking at sections of the genome of captive birds in which the nucleotides repeat multiple times, such as AGATAGATAGAT. Called microsatellites, these consecutively-repeating sections can be passed from parents to children, helping scientists identify parent-child and sibling-sibling relationships. “That allows you to make strategic decisions about how to pair (captive) individuals for mating...to avoid inbreeding,” Coleman says.
Jeffrey Coleman is studying the microsatellites inside the toucan genomes.
Courtesy Jeffrey Coleman
The alternative is to use a type of analysis that looks for a single DNA building block – a nucleotide – that differs in a given sequence. Called single nucleotide polymorphisms (SNPs, pronounced “snips”), they are very common and very accurate. Coleman says they are better than microsatellites for some uses. But scientists have already developed a large body of microsatellite data from multiple species, so microsatellites can shed more insights on relations.
Regardless of whether conservation programs use SNPs or microsatellites to guide captive breeding efforts, the goal is to help them build genetically diverse populations that eventually may supplement endangered populations in the wild. “The hope is that the ecosystem will be stable enough and that the populations (once reintroduced into the wild) will be able to survive and thrive,” says Coleman. History knows some good examples of captive breeding success.
The California condor, which had a total population of 27 in 1987, when the last wild birds were captured, is one of them. A captive breeding program boosted their numbers to 561 by the end of 2022. Of those, 347 of those are in the wild, according to the National Park Service.
Conservationists hope that their work on animals’ genetic diversity will help preserve and restore endangered species in captivity and the wild. DNA analysis is crucial to both types of efforts. The ability to apply genome sequencing to wildlife conservation brings a new level of accuracy that helps protect species and gives fresh insights that observation alone can’t provide.
“A lot of species are threatened,” Coleman says. “I hope this research will be a resource people can use to get more information on longer-term genealogies and different populations.”
DNA- and RNA-based electronic implants may revolutionize healthcare
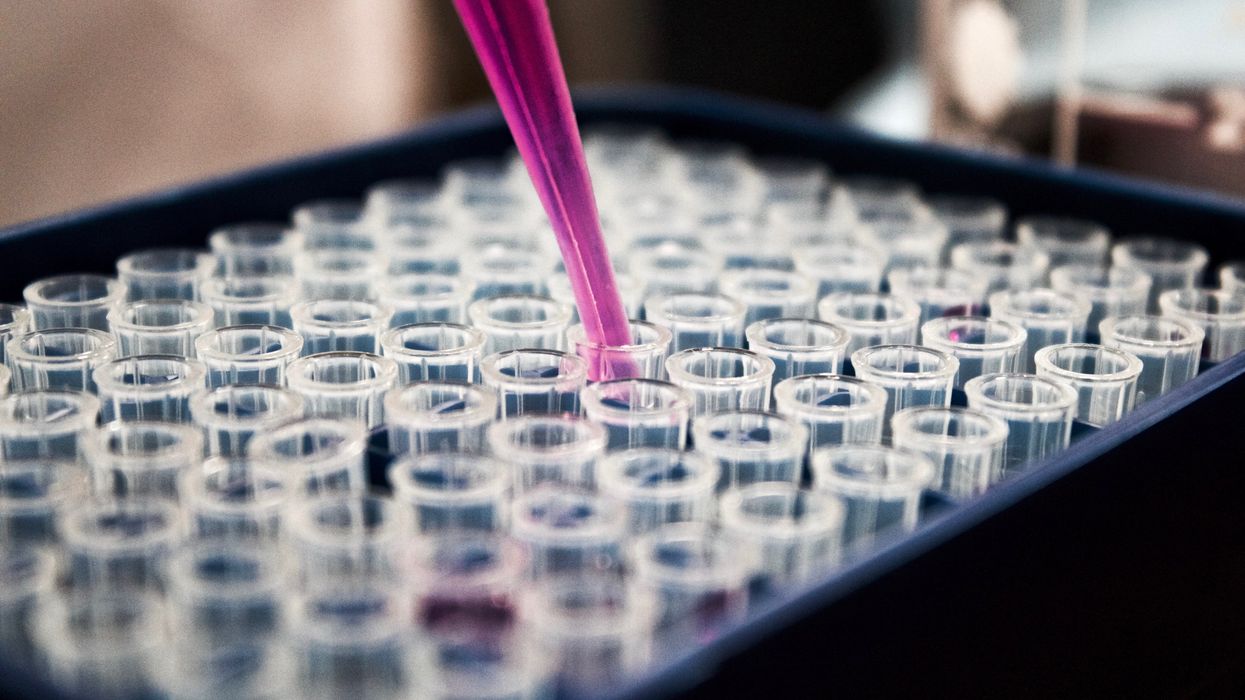
The test tubes contain tiny DNA/enzyme-based circuits, which comprise TRUMPET, a new type of electronic device, smaller than a cell.
Implantable electronic devices can significantly improve patients’ quality of life. A pacemaker can encourage the heart to beat more regularly. A neural implant, usually placed at the back of the skull, can help brain function and encourage higher neural activity. Current research on neural implants finds them helpful to patients with Parkinson’s disease, vision loss, hearing loss, and other nerve damage problems. Several of these implants, such as Elon Musk’s Neuralink, have already been approved by the FDA for human use.
Yet, pacemakers, neural implants, and other such electronic devices are not without problems. They require constant electricity, limited through batteries that need replacements. They also cause scarring. “The problem with doing this with electronics is that scar tissue forms,” explains Kate Adamala, an assistant professor of cell biology at the University of Minnesota Twin Cities. “Anytime you have something hard interacting with something soft [like muscle, skin, or tissue], the soft thing will scar. That's why there are no long-term neural implants right now.” To overcome these challenges, scientists are turning to biocomputing processes that use organic materials like DNA and RNA. Other promised benefits include “diagnostics and possibly therapeutic action, operating as nanorobots in living organisms,” writes Evgeny Katz, a professor of bioelectronics at Clarkson University, in his book DNA- And RNA-Based Computing Systems.
While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output.
Adamala’s research focuses on developing such biocomputing systems using DNA, RNA, proteins, and lipids. Using these molecules in the biocomputing systems allows the latter to be biocompatible with the human body, resulting in a natural healing process. In a recent Nature Communications study, Adamala and her team created a new biocomputing platform called TRUMPET (Transcriptional RNA Universal Multi-Purpose GatE PlaTform) which acts like a DNA-powered computer chip. “These biological systems can heal if you design them correctly,” adds Adamala. “So you can imagine a computer that will eventually heal itself.”
The basics of biocomputing
Biocomputing and regular computing have many similarities. Like regular computing, biocomputing works by running information through a series of gates, usually logic gates. A logic gate works as a fork in the road for an electronic circuit. The input will travel one way or another, giving two different outputs. An example logic gate is the AND gate, which has two inputs (A and B) and two different results. If both A and B are 1, the AND gate output will be 1. If only A is 1 and B is 0, the output will be 0 and vice versa. If both A and B are 0, the result will be 0. While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output. In this case, the DNA enters the logic gate as a single or double strand.
If the DNA is double-stranded, the system “digests” the DNA or destroys it, which results in non-fluorescence or “0” output. Conversely, if the DNA is single-stranded, it won’t be digested and instead will be copied by several enzymes in the biocomputing system, resulting in fluorescent RNA or a “1” output. And the output for this type of binary system can be expanded beyond fluorescence or not. For example, a “1” output might be the production of the enzyme insulin, while a “0” may be that no insulin is produced. “This kind of synergy between biology and computation is the essence of biocomputing,” says Stephanie Forrest, a professor and the director of the Biodesign Center for Biocomputing, Security and Society at Arizona State University.
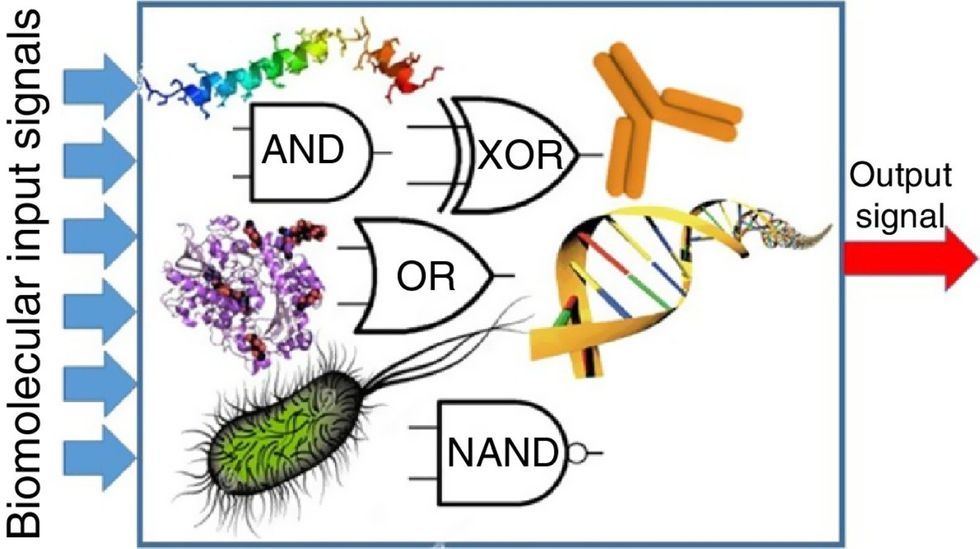
Biocomputing circles are made of DNA, RNA, proteins and even bacteria.
Evgeny Katz
The TRUMPET’s promise
Depending on whether the biocomputing system is placed directly inside a cell within the human body, or run in a test-tube, different environmental factors play a role. When an output is produced inside a cell, the cell's natural processes can amplify this output (for example, a specific protein or DNA strand), creating a solid signal. However, these cells can also be very leaky. “You want the cells to do the thing you ask them to do before they finish whatever their businesses, which is to grow, replicate, metabolize,” Adamala explains. “However, often the gate may be triggered without the right inputs, creating a false positive signal. So that's why natural logic gates are often leaky." While biocomputing outside a cell in a test tube can allow for tighter control over the logic gates, the outputs or signals cannot be amplified by a cell and are less potent.
TRUMPET, which is smaller than a cell, taps into both cellular and non-cellular biocomputing benefits. “At its core, it is a nonliving logic gate system,” Adamala states, “It's a DNA-based logic gate system. But because we use enzymes, and the readout is enzymatic [where an enzyme replicates the fluorescent RNA], we end up with signal amplification." This readout means that the output from the TRUMPET system, a fluorescent RNA strand, can be replicated by nearby enzymes in the platform, making the light signal stronger. "So it combines the best of both worlds,” Adamala adds.
These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body.
The TRUMPET biocomputing process is relatively straightforward. “If the DNA [input] shows up as single-stranded, it will not be digested [by the logic gate], and you get this nice fluorescent output as the RNA is made from the single-stranded DNA, and that's a 1,” Adamala explains. "And if the DNA input is double-stranded, it gets digested by the enzymes in the logic gate, and there is no RNA created from the DNA, so there is no fluorescence, and the output is 0." On the story's leading image above, if the tube is "lit" with a purple color, that is a binary 1 signal for computing. If it's "off" it is a 0.
While still in research, TRUMPET and other biocomputing systems promise significant benefits to personalized healthcare and medicine. These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body. The study’s lead author and graduate student Judee Sharon is already beginning to research TRUMPET's ability for earlier cancer diagnoses. Because the inputs for TRUMPET are single or double-stranded DNA, any mutated or cancerous DNA could theoretically be detected from the platform through the biocomputing process. Theoretically, devices like TRUMPET could be used to detect cancer and other diseases earlier.
Adamala sees TRUMPET not only as a detection system but also as a potential cancer drug delivery system. “Ideally, you would like the drug only to turn on when it senses the presence of a cancer cell. And that's how we use the logic gates, which work in response to inputs like cancerous DNA. Then the output can be the production of a small molecule or the release of a small molecule that can then go and kill what needs killing, in this case, a cancer cell. So we would like to develop applications that use this technology to control the logic gate response of a drug’s delivery to a cell.”
Although platforms like TRUMPET are making progress, a lot more work must be done before they can be used commercially. “The process of translating mechanisms and architecture from biology to computing and vice versa is still an art rather than a science,” says Forrest. “It requires deep computer science and biology knowledge,” she adds. “Some people have compared interdisciplinary science to fusion restaurants—not all combinations are successful, but when they are, the results are remarkable.”