New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
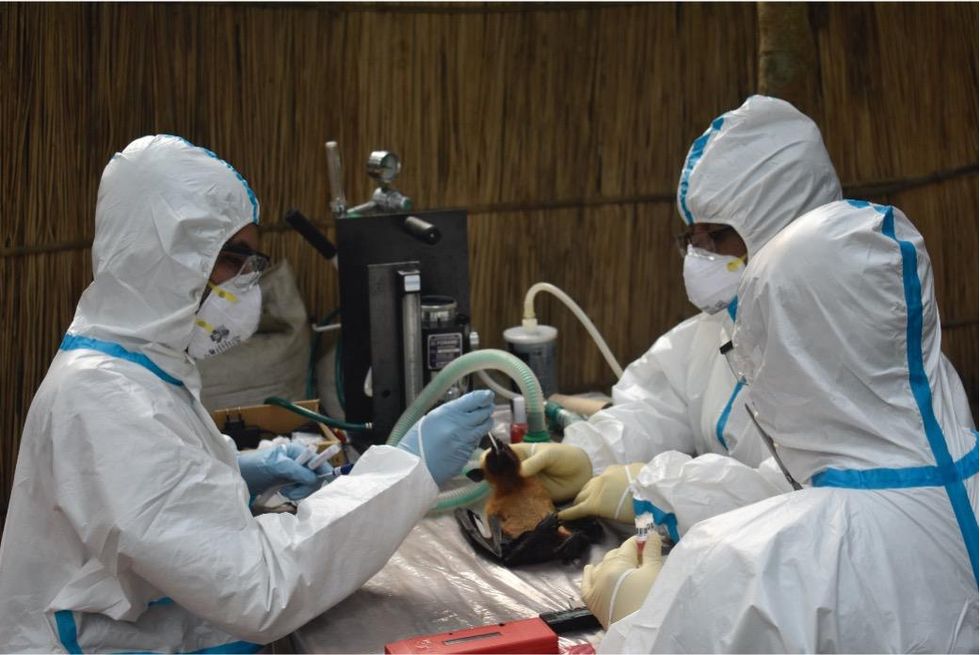
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
The CRISPR-Cas9 gene editing tool could be used to "turn off" pain directly, raising ethical questions for society.
Scientists have long been aware that some people live with what's known as "congenital insensitivity to pain"—the inability to register the tingles, jolts, and aches that alert most people to injury or illness.
"If you break the chain of transmission somewhere along there, it doesn't matter what the message is—the recipient will not get it."
On the ospposite end of the spectrum, others suffer from hyperalgesia, or extreme pain; for those with erythromelalgia, also known as "Man on Fire Syndrome," warm temperatures can feel like searing heat—even wearing socks and shoes can make walking unbearable.
Strangely enough, the two conditions can be traced to mutations in the same gene, SCN9A. It produces a protein that exists in spinal cells—specifically, in the dorsal root ganglion—which transmits the sensation of pain from the nerves at the peripheral site of an injury into the central nervous system and to the brain. This fact may become the key to pain relief for the roughly 20 percent of Americans who suffer from chronic pain, and countless other patients around the world.
"If you break the chain of transmission somewhere along there, it doesn't matter what the message is—the recipient will not get it," said Dr. Fyodor Urnov, director of the Innovative Genomics Institute and a professor of molecular and cell biology at the University of California, Berkeley. "For scientists and clinicians who study this, [there's] this consistent tracking of: You break this gene, you stop feeling pain; make this gene hyperactive, you feel lots of pain—that really cuts through the correlation versus causation question."
Researchers tried for years, without much success, to find a chemical that would block that protein from working and therefore mute the pain sensation. The CRISPR-Cas9 gene editing tool could completely sidestep that approach and "turn off" pain directly.
Yet as CRISPR makes such targeted therapies increasingly possible, the ethical questions surrounding gene editing have taken on a new and more urgent cast—particularly in light of the work of the disgraced Chinese scientist He Jiankui, who announced in late 2018 that he had created the world's first genetically edited babies. He used CRISPR to edit two embryos, with the goal of disabling a gene that makes people susceptible to HIV infection; but then took the unprecedented step of implanting the edited embryos for pregnancy and birth.
Edits to germline cells, like the ones He undertook, involve alterations to gametes or embryos and carry much higher risk than somatic cell edits, since changes will be passed on to any future generations. There are also concerns that imprecise edits could result in mutations and end up causing more disorders. Recent developments, particularly the "search-and replace" prime-editing technique published last fall, will help minimize those accidental edits, but the fact remains that we have little understanding of the long-term effects of these germline edits—for the future of the patients themselves, or for the broader gene pool.
"We need to have appropriate venues where we deliberate and consider the ethical, legal and social implications of gene editing as a society."
It is much harder to predict the effects, harmful or otherwise, on the larger human population as a result of interactions with the environment or other genetic variations; with somatic cell edits, on the other hand— like the ones that would be made in an individual to turn off pain—only the person receiving the treatment is affected.
Beyond the somatic/germline distinction, there is also a larger ethical question over how much genetic interference society is willing to tolerate, which may be couched as the difference between therapeutic editing—interventions in response to a demonstrated medical need—and "enhancement" editing. The Chinese scientist He was roundly criticized in the scientific community for the fact that there are already much safer and more proven methods of preventing the parent-to-child transmission of HIV through the IVF process, making his genetic edits medically unnecessary. (The edits may also have increased the girls' risk of susceptibility to other viruses, like influenza and the West Nile virus.)
Yet there are even more extreme goals that CRISPR could be used to reach, ones further removed from any sort of medical treatment. The 1997 science fiction movie Gattaca imagined a dystopian future where genetic selection for strength and intelligence is common, creating a society that explicitly and unapologetically endorses eugenics. In the real world, Russian President Vladimir Putin has commented that genetic editing could be used to create "a genius mathematician, a brilliant musician or a soldier, a man who can fight without fear, compassion, regret or pain."
"[Such uses] would be considered using gene editing for 'enhancement,'" said Dr. Zubin Master, an associate professor of biomedical ethics at the Mayo Clinic, who noted that a series of studies have strongly suggested that members of the public, in the U.S. and around the world, are much less amenable to the prospect of gene editing for these purposes than for the treatment of illness and disease.
Putin's comments were made in 2017, before news of He's experiment broke; since then no country has moved to continue experiments on germline editing (although one Russian IVF specialist, Denis Rebrikov, appears ready to do so, if given approval). Master noted that the World Health Organization has an 18-person committee currently dedicated to considering these questions. The Expert Advisory Committee on Developing Global Standards for Governance and Oversight of Human Genome Editing first convened in March 2019; that July, it issued a recommendation to regulatory and ethics authorities in all countries to refrain from approving clinical application requests for work on human germline genome editing—the kind of alterations to genetic cells used by He. The committee's report and a fleshed-out set of guidelines is expected after its final meeting, in Geneva this September (unless the COVID-19 pandemic disrupts the timeline).
Regardless of the WHO's report, in the U.S., all regulations of new medical procedures are overseen at the federal level, subjected to extensive regulatory review by the FDA; the chance of any doctor or company going rogue is minimal to none. Likewise, the challenges we face are more on the regulatory end of the spectrum than the Gattaca end. Dr. Stephanie Malia Fullerton, a bioethics professor at the University of Washington, pointed out that eugenics not only typically involves state-sponsored control of reproduction, but requires a much more clearly delineated genetic basis of common complex traits—indeed, SCN9A is one way to get to pain, but is not the only source—and suggested that current concerns about over-prescribing opioids are a more pressing question for society to address.
In fact, Navega Therapeutics, based in San Diego, hopes to find out whether the intersection of this research into SCN9A and CRISPR would be an effective way to address the U.S. opioid crisis. Currently in a preclinical funding stage, Navega's approach focuses on editing epigenetic molecules attached to the basic DNA strand—the idea is that the gene's expression can be activated or suppressed rather than removed entirely, reducing the risk of unwanted side effects from permanently altering the genetic code.
As these studies focused on the sensation of pain go forward, what we are likely to see simultaneously is the use of CRISPR to target diseases that are the root causes of that pain. Last summer, Victoria Gray, a Mississippi woman with sickle cell disease was the second-ever person to be treated with CRISPR therapy in the U.S. The disease is caused by a genetic mutation that creates malformed blood cells, which can't carry oxygen as normal and get stuck inside blood vessels, causing debilitating pain. For the study, conducted in concert with CRISPR Therapeutics, of Cambridge, Mass., cells were removed from Gray's bone marrow, modified using CRISPR, and infused back into her body, a technique called ex vivo editing.
In early February this year, researchers at the University of Pennsylvania published a study on a first-in-human phase 1 clinical trial, in which three patients with advanced cancer received an infusion of ex vivo engineered T cells in an effort to improve antitumor immunity. The modified cells persisted for up to nine months, and the patients experienced no serious adverse side effects, suggesting that this sort of therapeutic gene editing can be performed safely and could potentially allow patients to avoid the excruciating process of chemotherapy.
Then, just this spring, researchers made another advance: The first attempt at in vivo CRISPR editing—where the edits happen inside the patient's body—is currently underway, as doctors attempt to treat a patient blinded by Leber congenital amaurosis, a rare genetic disorder. In an Oregon study sponsored by Editas Medicine and Allergan, the patient, a volunteer, was injected with a harmless virus carrying CRISPR gene-editing machinery; the hope is that the tool will be able to edit out the genetic defect and restore production of a crucial protein. Based on preliminary safety reports, the study has been cleared to continue, and data on higher doses may be available by the end of 2020. Editas Medicine and CRISPR Therapeutics are joined in this sphere by Intellia Therapeutics, which is seeking approval for a trial later this year on amyloidosis, a rare liver condition.
For any such treatment targeting SCN9A to make its way to human subjects, it would first need to undergo years' worth of testing—on mice, on primates, and then on volunteer patients after an extended informed-consent process. If everything went perfectly, Urnov estimates it could take at least three to four years end to end and cost between $5 and 10 million—but that "if" is huge.
"The idea of a regular human being, genetically pure of pain?"
And as that happens, "we need to have appropriate venues where we deliberate and consider the ethical, legal and social implications of gene editing as a society," Master said. CRISPR itself is open-source, but its application is subject to the approval of governments, institutions, and societies, which will need to figure out where to draw the line between miracle treatments and playing God. Something as unpleasant and ubiquitous as pain may in fact be the most appropriate place to start.
"The pain circuit is very old," Urnov said. "We have evolved with the senses that we have, and have become the species that we are, as a result of who we are, physiologically. Yes, I take Advil—but when I get a headache! The idea of a regular human being, genetically pure of pain?... The permanent disabling or turning down of the pain sensation, for anything other than a medical reason? … That seems to be challenging Mother Nature in the wrong ways."
A biosensor in development (inset) could potentially be used to detect novel viruses in transit hubs like Grand Central Station in New York City.
The unprecedented scale and impact of the COVID-19 pandemic has caused scientists and engineers around the world to stop whatever they were working on and shift their research toward understanding a novel virus instead.
"We have confidence that we can use our system in the next pandemic."
For Guangyu Qiu, normally an environmental engineer at the Swiss Federal Laboratories for Materials Science and Technology, that means finding a clever way to take his work on detecting pollution in the air and apply it to living pathogens instead. He's developing a new type of biosensor to make disease diagnostics and detection faster and more accurate than what's currently available.
But even though this pandemic was the impetus for designing a new biosensor, Qiu actually has his eye on future disease outbreaks. He admits that it's unlikely his device will play a role in quelling this virus, but says researchers already need to be thinking about how to make better tools to fight the next one — because there will be a next one.
"In the last 20 years, there [have been] three different coronavirus [outbreaks] ... so we have to prepare for the coming one," Qiu says. "We have confidence that we can use our system in the next pandemic."
"A Really, Really Neat Idea"
His main concern is the diagnostic tool that's currently front and center for testing patients for SARS-Cov-2, the virus causing the novel coronavirus disease. The tool, called PCR (short for reverse transcription polymerase chain reaction), is the gold standard because it excels at detecting viruses in even very small samples of mucus. PCR can amplify genetic material in the limited sample and look for a genetic code matching the virus in question. But in many parts of the world, mucus samples have to be sent out to laboratories for that work, and results can take days to return. PCR is also notoriously prone to false positives and negatives.
"I read a lot of newspapers that report[ed] ... a lot of false negative or false positive results at the very beginning of the outbreak," Qiu says. "It's not good for protecting people to prevent further transmission of the disease."
So he set out to build a more sensitive device—one that's less likely to give you a false result. Qiu's biosensor relies on an idea similar to the dual-factor authentication required of anyone trying to access a secure webpage. Instead of verifying that a virus is really present by using one way of detecting genetic code, as with PCR, this biosensor asks for two forms of ID.
SARS-CoV-2 is what's called an RNA virus, which means it has a single strand of genetic code, unlike double-stranded DNA. Inside Qiu's biosensor are receptors with the complementary code for this particular virus' RNA; if the virus is present, its RNA will bind with the receptors, locking together like velcro. The biosensor also contains a prism and a laser that work together to verify that this RNA really belongs to SARS-CoV-2 by looking for a specific wavelength of light and temperature.
If the biosensor doesn't detect either, or only registers a match for one and not the other, then it can't produce a positive result. This multi-step authentication process helps make sure that the RNA binding with the receptors isn't a genetically similar coronavirus like SARS-CoV, known for its 2003 outbreak, or MERS-CoV, which caused an epidemic in 2012.
It could also be fitted to detect future novel viruses once their genomes are sequenced.
The dual-feature design of this biosensor "is a really, really neat idea that I have not seen before with other sensor technology," says Erin Bromage, a professor of infection and immunology at the University of Massachusetts Dartmouth; he was not involved in designing or testing Qiu's biosensor. "It makes you feel more secure that when you have a positive, you've really got a positive."
The light and temperature sensors are not in themselves new inventions, but the combination is a first. The part of the device that uses light to detect particles is actually central to Qiu's normal stream of environmental research, and is a versatile tool he's been working with for a long time to detect aerosols in the atmosphere and heavy metals in drinking water.
Bromage says this is a plus. "It's not high-risk in the sense that how they do this is unique, or not validated. They've taken aspects of really proven technology and sort of combined it together."
This new biosensor is still a prototype that will take at least another 12 months to validate in real world scenarios, though. The device is sound from a biological perspective and is sensitive enough to reliably detect SARS-CoV-2 — and to not be tricked by genetically similar viruses like SARS-CoV — but there is still a lot of engineering work that needs to be done in order for it to work outside the lab. Qiu says it's unlikely that the sensor will help minimize the impact of this pandemic, but the RNA receptors, prism, and laser inside the device can be customized to detect other viruses that may crop up in the future.
"If we choose another sequence—like SARS, like MERS, or like normal seasonal flu—we can detect other viruses, or even bacteria," Qiu says. "This device is very flexible."
It could also be fitted to detect future novel viruses once their genomes are sequenced.
The Long-Term Vision: Hospitals and Transit Hubs
The device has been designed to connect with two other systems: an air sampler and a microprocessor because the goal is to make it portable, and able to pick up samples from the air in hospitals or public areas like train stations or airports. A virus could hopefully be detected before it silently spreads and erupts into another global pandemic. In the case of SARS-CoV-2, there has been conflicting research about whether or not the virus is truly airborne (though it can be spread by droplets that briefly move through the air after a cough or sneeze), whereas the highly contagious RNA virus that causes measles can remain in the air for up to two hours.
"They've got a lot on the front end to work out," Bromage says. "They've got to work out how to capture and concentrate a virus, extract the RNA from the virus, and then get it onto the sensor. That's some pretty big hurdles, and may take some engineering that doesn't exist right now. But, if they can do that, then that works out really quite well."
One of the major obstacles in containing the COVID-19 pandemic has been in deploying accurate, quick tools that can be used for early detection of a virus outbreak and for later tracing its spread. That will still be true the next time a novel virus rears its head, and it's why Qiu feels that even if his biosensor can't help just yet, the research is still worth the effort.
It could also be fitted to detect future novel viruses once their genomes are sequenced.
The dual-feature design of this biosensor "is a really, really neat idea that I have not seen before with other sensor technology," says Erin Bromage, a professor of infection and immunology at the University of Massachusetts Dartmouth; he was not involved in designing or testing Qiu's biosensor. "It makes you feel more secure that when you have a positive, you've really got a positive."
The light and temperature sensors are not in themselves new inventions, but the combination is a first. The part of the device that uses light to detect particles is actually central to Qiu's normal stream of environmental research, and is a versatile tool he's been working with for a long time to detect aerosols in the atmosphere and heavy metals in drinking water.
Bromage says this is a plus. "It's not high-risk in the sense that how they do this is unique, or not validated. They've taken aspects of really proven technology and sort of combined it together."
This new biosensor is still a prototype that will take at least another 12 months to validate in real world scenarios, though. The device is sound from a biological perspective and is sensitive enough to reliably detect SARS-CoV-2 — and to not be tricked by genetically similar viruses like SARS-CoV — but there is still a lot of engineering work that needs to be done in order for it to work outside the lab. Qiu says it's unlikely that the sensor will help minimize the impact of this pandemic, but the RNA receptors, prism, and laser inside the device can be customized to detect other viruses that may crop up in the future.
"If we choose another sequence—like SARS, like MERS, or like normal seasonal flu—we can detect other viruses, or even bacteria," Qiu says. "This device is very flexible."
It could also be fitted to detect future novel viruses once their genomes are sequenced.
The Long-Term Vision: Hospitals and Transit Hubs
The device has been designed to connect with two other systems: an air sampler and a microprocessor because the goal is to make it portable, and able to pick up samples from the air in hospitals or public areas like train stations or airports. A virus could hopefully be detected before it silently spreads and erupts into another global pandemic. In the case of SARS-CoV-2, there has been conflicting research about whether or not the virus is truly airborne (though it can be spread by droplets that briefly move through the air after a cough or sneeze), whereas the highly contagious RNA virus that causes measles can remain in the air for up to two hours.
"They've got a lot on the front end to work out," Bromage says. "They've got to work out how to capture and concentrate a virus, extract the RNA from the virus, and then get it onto the sensor. That's some pretty big hurdles, and may take some engineering that doesn't exist right now. But, if they can do that, then that works out really quite well."
One of the major obstacles in containing the COVID-19 pandemic has been in deploying accurate, quick tools that can be used for early detection of a virus outbreak and for later tracing its spread. That will still be true the next time a novel virus rears its head, and it's why Qiu feels that even if his biosensor can't help just yet, the research is still worth the effort.

