Medical Breakthroughs Set to be Fast-Tracked by Innovative New Health Agency

Pippy Rogers, second from left, with her four siblings, who worry that they are at risk for Alzheimer's and are calling for an acceleration of research.
In 2007, Matthew Might's son, Bertrand, was born with a life-threatening disease that was so rare, doctors couldn't diagnose it. Might, a computer scientist and biologist, eventually realized, "Oh my gosh, he's the only patient in the world with this disease right now." To find effective treatments, new methodologies would need to be developed. But there was no process or playbook for doing that.
Might took it upon himself, along with a team of specialists, to try to find a cure. "What Bertrand really taught me was the visceral sense of urgency when there's suffering, and how to act on that," he said.
He calls it "the agency of urgency"—and patients with more common diseases, such as cancer and Alzheimer's, often feel that same need to take matters into their own hands, as they find their hopes for new treatments running up against bureaucratic systems designed to advance in small, steady steps, not leaps and bounds. "We all hope for a cure," said Florence "Pippy" Rogers, a 65-year-old volunteer with Georgia's chapter of the Alzheimer's Association. She lost her mother to the disease and, these days, worries about herself and her four siblings. "We need to keep accelerating research."
We have a fresh example of what can be achieved by fast-tracking discoveries in healthcare: Covid-19 vaccines.
President Biden has pushed for cancer moonshots since the disease took the life of his son, Beau, in 2015. His administration has now requested $6.5 billion to start a new agency in 2022, called the Advanced Research Projects Agency for Health, or ARPA-H, within the National Institutes of Health. It's based on DARPA, the Department of Defense agency known for hatching world-changing technologies such as drones, GPS and ARPANET, which became the internet.
We have a fresh example of what can be achieved by fast-tracking discoveries in healthcare: Covid-19 vaccines. "Operation Warp Speed was using ARPA-like principles," said Might. "It showed that in a moment of crisis, institutions like NIH can think in an ARPA-like way. So now the question is, why don't we do that all the time?"
But applying the DARPA model to health involves several challenging decisions. I asked experts what could be the hardest question facing advocates of ARPA-H: which health problems it should seek to address. "All the wonderful choices lead to the problem of which ones to choose and prioritize," said Sudip Parikh, CEO of the American Association for the Advancement of Science and executive publisher of the Science family of journals. "There is no objectively right answer."
The Agency of Urgency
ARPA-H will borrow at least three critical ingredients from DARPA: goal-oriented project managers, many from industry; aggressive public-private partnerships; and collaboration among fields that don't always interact. The DARPA concept has been applied to other purposes, including energy and homeland security, with promising results. "We're learning that 'ARPA-ism' is a franchisable model," said Might, a former principal investigator on DARPA projects.
The federal government already pours billions of dollars into advancing research on life-threatening diseases, with much of it channeled through the National Institutes of Health. But the purpose of ARPA-H "isn't just the usual suspects that NIH would fund," said David Walt, a Harvard biochemist, an innovator in gene sequencing and former chair of DARPA's Defense Science Research Council. Whereas some NIH-funded studies aim to gradually improve our understanding of diseases, ARPA-H projects will give full focus to real-world applications; they'll use essential findings from NIH research as starting points, drawing from them to rapidly engineer new technologies that could save lives.
And, ultimately, billions in healthcare costs, if ARPA-H lives up to its predecessor's track record; DARPA's breakthroughs have been economic game-changers, while its fail-fast approach—quickly pulling the plug on projects that aren't panning out—helps to avoid sunken costs. ARPA-H could fuel activities similar to the human genome project, which used existing research to map the base pairs that make up DNA, opening new doors for the biotech industry, sparking economic growth and creating hundreds of thousands of new jobs.
Despite a nearly $4 trillion health economy, "we aren't innovating when it comes to technological capabilities for health," said Liz Feld, president of the Suzanne Wright Foundation for pancreatic cancer.
Individual Diseases Ripe for Innovation
Although the need for innovation is clear, which diseases ARPA-H should tackle is less apparent. One important consideration when choosing health priorities could be "how many people suffer from a disease," said Nancy Kass, a professor of bioethics and public health at Johns Hopkins.
That perspective could justify cancer as a top objective. Cancer and heart disease have long been the two major killers in the U.S. Leonidas Platanias, professor of oncology at Northwestern and director of its cancer center, noted that we've already made significant progress on heart disease. "Anti-cholesterol drugs really have a wide impact," he said. "I don't want to compare one disease to another, but I think cancer may be the most challenging. We need even bigger breakthroughs." He wondered whether ARPA-H should be linked to the part of NIH dedicated to cancer, the National Cancer Institute, "to take maximum advantage of what happens" there.
Previous cancer moonshots have laid a foundation for success. And this sort of disease-by-disease approach makes sense in a way. "We know that concentrating on some diseases has led to treatments," said Parikh. "Think of spinal muscular atrophy or cystic fibrosis. Now, imagine if immune therapies were discovered ten years earlier."
But many advocates think ARPA-H should choose projects that don't revolve around any one disease. "It absolutely has to be disease agnostic," said Feld, president of the pancreatic cancer foundation. "We cannot reach ARPA-H's potential if it's subject to the advocacy of individual patient groups who think their disease is worse than the guy's disease next to them. That's not the way the DARPA model works." Platanias agreed that ARPA-H should "pick the highest concepts and developments that have the best chance" of success.
Finding Connections Between Diseases
Kass, the Hopkins bioethicist, believes that ARPA-H should walk a balance, with some projects focusing on specific diseases and others aspiring to solutions with broader applications, spanning multiple diseases. Being impartial, some have noted, might involve looking at the total "life years" saved by a health innovation; the more diseases addressed by a given breakthrough, the more years of healthy living it may confer. The social and economic value should increase as well.
For multiple payoffs, ARPA-H could concentrate on rare diseases, which can yield important insights for many other diseases, said Might. Every case of cancer and Alzheimer's is, in a way, its own rare disease. Cancer is a genetic disease, like his son Bertrand's rare disorder, and mutations vary widely across cancer patients. "It's safe to say that no two people have ever actually had the same cancer," said Might. In theory, solutions for rare diseases could help us understand how to individualize treatments for more common diseases.
Many experts I talked with support another priority for ARPA-H with implications for multiple diseases: therapies that slow down the aging process. "Aging is the greatest risk factor for every major disease that NIH is studying," said Matt Kaeberlein, a bio-gerontologist at the University of Washington. Yet, "half of one percent of the NIH budget goes to researching the biology of aging. An ARPA-H sized budget would push the field forward at a pace that's hard to imagine."
Might agreed. "It could take ARPA-H to get past the weird stigmas around aging-related research. It could have a tremendous impact on the field."
For example, ARPA-H could try to use mRNA technology to express proteins that affect biological aging, said Kaeberlein. It's an engineering project well-suited to the DARPA model. So is harnessing machine learning to identify biomarkers that assess how fast people are aging. Biological aging clocks, if validated, could quickly reveal whether proposed therapies for aging are working or not. "I think there's huge value in that," said Kaeberlein.
By delivering breakthroughs in computation, ARPA-H could improve diagnostics for many different diseases. That could include improving biowearables for continuously monitoring blood pressure—a hypothetical mentioned in the White House's concept paper on ARPA-H—and advanced imaging technologies. "The high cost of medical imaging is a leading reason why our healthcare costs are the highest in the world," said Feld. "There's no detection test for ALS. No brain detection for Alzheimer's. Innovations in detection technology would save on cost and human suffering."
Some biotech companies may be skeptical about the financial rewards of accelerating such technologies. But ARPA-H could fund public-private partnerships to "de-risk" biotech's involvement—an incentive that harkens back to the advance purchase contracts that companies got during Covid. (Some groups have suggested that ARPA-H could provide advance purchase agreements.)
Parikh is less bullish on creating diagnostics through ARPA-H. Like DARPA, Biden's health agency will enjoy some independence from federal oversight; it may even be located hundreds of miles from DC. That freedom affords some breathing room for innovation, but it could also make it tougher to ensure that algorithms fully consider diverse populations. "That part I really would like the government more involved in," Parikh said.
Might thinks ARPA-H should also explore innovations in clinical trials, which many patients and medical communities view as grindingly slow and requiring too many participants. "We can approve drugs for very tiny patient populations, even at the level of the individual," he said, while emphasizing the need for safety. But Platanias thinks the FDA has become much more flexible in recent years. In the cancer field, at least, "You now see faster approvals for more drugs. Having [more] shortcuts on clinical trial approvals is not necessarily a good idea."
With so many options on the table, ARPA-H needs to show the public a clear framework for measuring the value of potential projects. Kass warned that well-resourced advocates could skew the agency's priorities. They've affected health outcomes before, she noted; fundraising may partly explain larger increases in life expectancy for cystic fibrosis than sickle cell anemia. Engaging diverse communities is a must for ARPA-H. So are partnerships to get the agency's outputs to people who need them. "Research is half the equation," said Kass. "If we don't ensure implementation and access, who cares." The White House concept paper on ARPA-H made a similar point.
As Congress works on authorizing ARPA-H this year, Might is doing what he can to ensure better access to innovation on a patient-by-patient basis. Last year, his son, Bertrand, passed away suddenly from his disorder. He was 12. But Might's sense of urgency has persisted, as he directs the Precision Medicine Institute at the University of Alabama-Birmingham. That urgency "can be carried into an agency like ARPA-H," he said. "It guides what I do as I apply for funding, because I'm trying to build the infrastructure that other parents need. So they don't have to build it from scratch like I did."
DNA- and RNA-based electronic implants may revolutionize healthcare
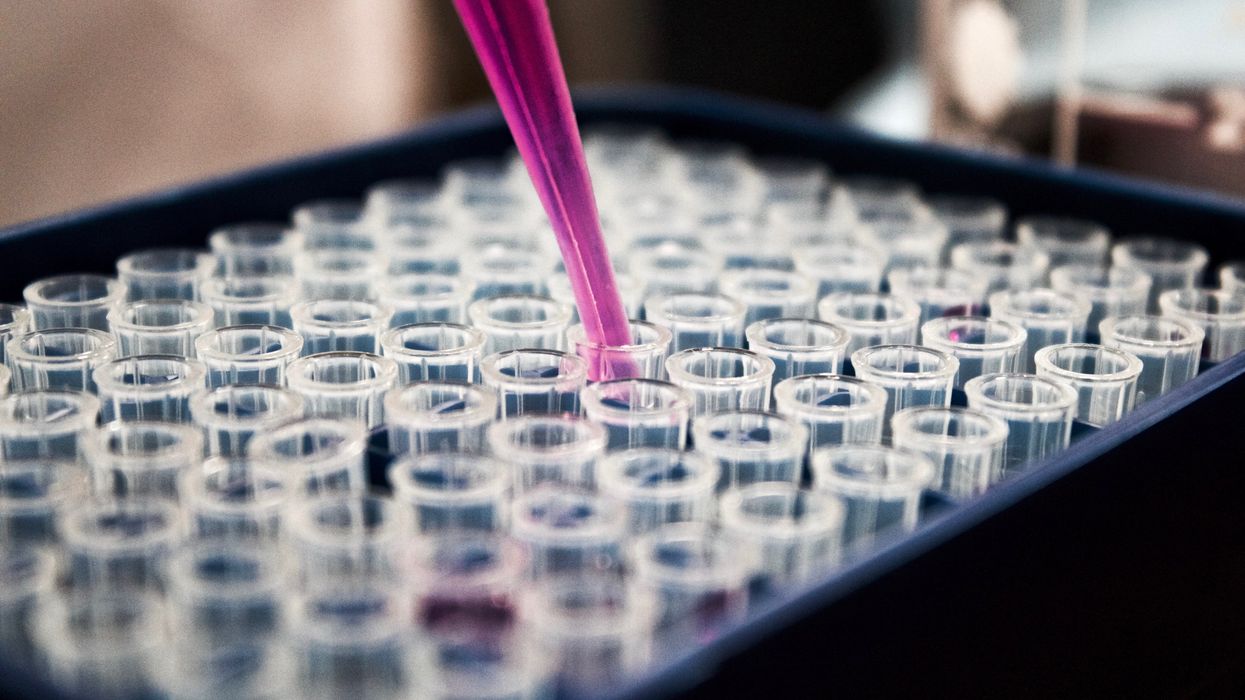
The test tubes contain tiny DNA/enzyme-based circuits, which comprise TRUMPET, a new type of electronic device, smaller than a cell.
Implantable electronic devices can significantly improve patients’ quality of life. A pacemaker can encourage the heart to beat more regularly. A neural implant, usually placed at the back of the skull, can help brain function and encourage higher neural activity. Current research on neural implants finds them helpful to patients with Parkinson’s disease, vision loss, hearing loss, and other nerve damage problems. Several of these implants, such as Elon Musk’s Neuralink, have already been approved by the FDA for human use.
Yet, pacemakers, neural implants, and other such electronic devices are not without problems. They require constant electricity, limited through batteries that need replacements. They also cause scarring. “The problem with doing this with electronics is that scar tissue forms,” explains Kate Adamala, an assistant professor of cell biology at the University of Minnesota Twin Cities. “Anytime you have something hard interacting with something soft [like muscle, skin, or tissue], the soft thing will scar. That's why there are no long-term neural implants right now.” To overcome these challenges, scientists are turning to biocomputing processes that use organic materials like DNA and RNA. Other promised benefits include “diagnostics and possibly therapeutic action, operating as nanorobots in living organisms,” writes Evgeny Katz, a professor of bioelectronics at Clarkson University, in his book DNA- And RNA-Based Computing Systems.
While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output.
Adamala’s research focuses on developing such biocomputing systems using DNA, RNA, proteins, and lipids. Using these molecules in the biocomputing systems allows the latter to be biocompatible with the human body, resulting in a natural healing process. In a recent Nature Communications study, Adamala and her team created a new biocomputing platform called TRUMPET (Transcriptional RNA Universal Multi-Purpose GatE PlaTform) which acts like a DNA-powered computer chip. “These biological systems can heal if you design them correctly,” adds Adamala. “So you can imagine a computer that will eventually heal itself.”
The basics of biocomputing
Biocomputing and regular computing have many similarities. Like regular computing, biocomputing works by running information through a series of gates, usually logic gates. A logic gate works as a fork in the road for an electronic circuit. The input will travel one way or another, giving two different outputs. An example logic gate is the AND gate, which has two inputs (A and B) and two different results. If both A and B are 1, the AND gate output will be 1. If only A is 1 and B is 0, the output will be 0 and vice versa. If both A and B are 0, the result will be 0. While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output. In this case, the DNA enters the logic gate as a single or double strand.
If the DNA is double-stranded, the system “digests” the DNA or destroys it, which results in non-fluorescence or “0” output. Conversely, if the DNA is single-stranded, it won’t be digested and instead will be copied by several enzymes in the biocomputing system, resulting in fluorescent RNA or a “1” output. And the output for this type of binary system can be expanded beyond fluorescence or not. For example, a “1” output might be the production of the enzyme insulin, while a “0” may be that no insulin is produced. “This kind of synergy between biology and computation is the essence of biocomputing,” says Stephanie Forrest, a professor and the director of the Biodesign Center for Biocomputing, Security and Society at Arizona State University.
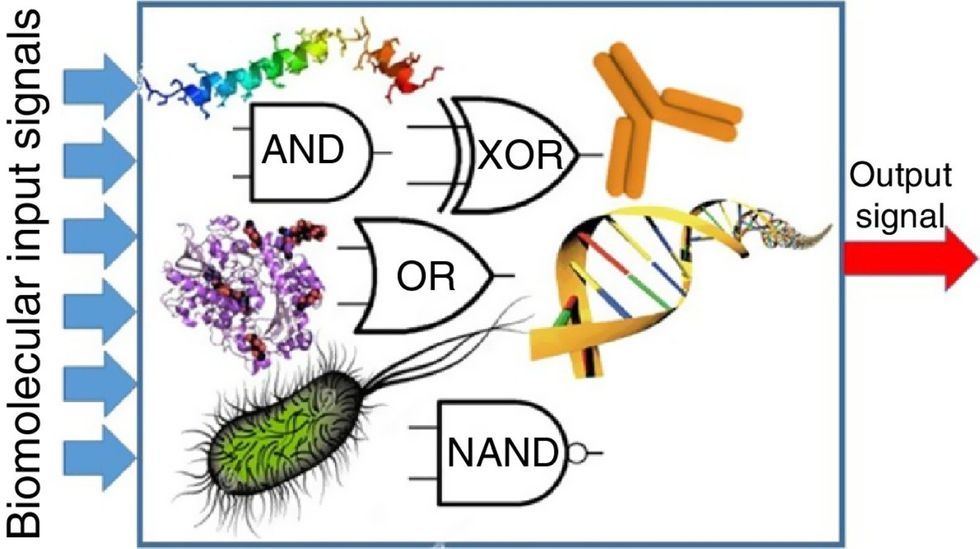
Biocomputing circles are made of DNA, RNA, proteins and even bacteria.
Evgeny Katz
The TRUMPET’s promise
Depending on whether the biocomputing system is placed directly inside a cell within the human body, or run in a test-tube, different environmental factors play a role. When an output is produced inside a cell, the cell's natural processes can amplify this output (for example, a specific protein or DNA strand), creating a solid signal. However, these cells can also be very leaky. “You want the cells to do the thing you ask them to do before they finish whatever their businesses, which is to grow, replicate, metabolize,” Adamala explains. “However, often the gate may be triggered without the right inputs, creating a false positive signal. So that's why natural logic gates are often leaky." While biocomputing outside a cell in a test tube can allow for tighter control over the logic gates, the outputs or signals cannot be amplified by a cell and are less potent.
TRUMPET, which is smaller than a cell, taps into both cellular and non-cellular biocomputing benefits. “At its core, it is a nonliving logic gate system,” Adamala states, “It's a DNA-based logic gate system. But because we use enzymes, and the readout is enzymatic [where an enzyme replicates the fluorescent RNA], we end up with signal amplification." This readout means that the output from the TRUMPET system, a fluorescent RNA strand, can be replicated by nearby enzymes in the platform, making the light signal stronger. "So it combines the best of both worlds,” Adamala adds.
These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body.
The TRUMPET biocomputing process is relatively straightforward. “If the DNA [input] shows up as single-stranded, it will not be digested [by the logic gate], and you get this nice fluorescent output as the RNA is made from the single-stranded DNA, and that's a 1,” Adamala explains. "And if the DNA input is double-stranded, it gets digested by the enzymes in the logic gate, and there is no RNA created from the DNA, so there is no fluorescence, and the output is 0." On the story's leading image above, if the tube is "lit" with a purple color, that is a binary 1 signal for computing. If it's "off" it is a 0.
While still in research, TRUMPET and other biocomputing systems promise significant benefits to personalized healthcare and medicine. These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body. The study’s lead author and graduate student Judee Sharon is already beginning to research TRUMPET's ability for earlier cancer diagnoses. Because the inputs for TRUMPET are single or double-stranded DNA, any mutated or cancerous DNA could theoretically be detected from the platform through the biocomputing process. Theoretically, devices like TRUMPET could be used to detect cancer and other diseases earlier.
Adamala sees TRUMPET not only as a detection system but also as a potential cancer drug delivery system. “Ideally, you would like the drug only to turn on when it senses the presence of a cancer cell. And that's how we use the logic gates, which work in response to inputs like cancerous DNA. Then the output can be the production of a small molecule or the release of a small molecule that can then go and kill what needs killing, in this case, a cancer cell. So we would like to develop applications that use this technology to control the logic gate response of a drug’s delivery to a cell.”
Although platforms like TRUMPET are making progress, a lot more work must be done before they can be used commercially. “The process of translating mechanisms and architecture from biology to computing and vice versa is still an art rather than a science,” says Forrest. “It requires deep computer science and biology knowledge,” she adds. “Some people have compared interdisciplinary science to fusion restaurants—not all combinations are successful, but when they are, the results are remarkable.”
Crickets are low on fat, high on protein, and can be farmed sustainably. They are also crunchy.
In today’s podcast episode, Leaps.org Deputy Editor Lina Zeldovich speaks about the health and ecological benefits of farming crickets for human consumption with Bicky Nguyen, who joins Lina from Vietnam. Bicky and her business partner Nam Dang operate an insect farm named CricketOne. Motivated by the idea of sustainable and healthy protein production, they started their unconventional endeavor a few years ago, despite numerous naysayers who didn’t believe that humans would ever consider munching on bugs.
Yet, making creepy crawlers part of our diet offers many health and planetary advantages. Food production needs to match the rise in global population, estimated to reach 10 billion by 2050. One challenge is that some of our current practices are inefficient, polluting and wasteful. According to nonprofit EarthSave.org, it takes 2,500 gallons of water, 12 pounds of grain, 35 pounds of topsoil and the energy equivalent of one gallon of gasoline to produce one pound of feedlot beef, although exact statistics vary between sources.
Meanwhile, insects are easy to grow, high on protein and low on fat. When roasted with salt, they make crunchy snacks. When chopped up, they transform into delicious pâtes, says Bicky, who invents her own cricket recipes and serves them at industry and public events. Maybe that’s why some research predicts that edible insects market may grow to almost $10 billion by 2030. Tune in for a delectable chat on this alternative and sustainable protein.
Listen on Apple | Listen on Spotify | Listen on Stitcher | Listen on Amazon | Listen on Google
Further reading:
More info on Bicky Nguyen
https://yseali.fulbright.edu.vn/en/faculty/bicky-n...
The environmental footprint of beef production
https://www.earthsave.org/environment.htm
https://www.watercalculator.org/news/articles/beef-king-big-water-footprints/
https://www.frontiersin.org/articles/10.3389/fsufs.2019.00005/full
https://ourworldindata.org/carbon-footprint-food-methane
Insect farming as a source of sustainable protein
https://www.insectgourmet.com/insect-farming-growing-bugs-for-protein/
https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/insect-farming
Cricket flour is taking the world by storm
https://www.cricketflours.com/
https://talk-commerce.com/blog/what-brands-use-cricket-flour-and-why/

Lina Zeldovich has written about science, medicine and technology for Popular Science, Smithsonian, National Geographic, Scientific American, Reader’s Digest, the New York Times and other major national and international publications. A Columbia J-School alumna, she has won several awards for her stories, including the ASJA Crisis Coverage Award for Covid reporting, and has been a contributing editor at Nautilus Magazine. In 2021, Zeldovich released her first book, The Other Dark Matter, published by the University of Chicago Press, about the science and business of turning waste into wealth and health. You can find her on http://linazeldovich.com/ and @linazeldovich.